PathoGenoMics ERA-NET

FCT is a partner in the PathoGenoMics (PGM) network, an ERA-NET that began in 2004 and involved 15 partners from 10 European countries (Portugal, Germany, Austria, Finland, France, Hungary, Israel, Latvia, Portugal, Slovenia, and Spain), which cooperated to promote and fund transnational research on the genomics of microorganisms that cause disease in humans (with a special emphasis on genomic studies of bacteria and fungi) and to implement the European Research Area (ERA).
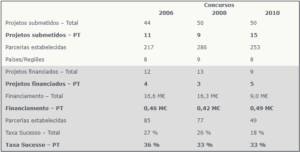
The PGM launched three Calls (2006, 2008, and 2010) with a total budget of €46 million. Part of the budget available for the Calls was dedicated to consortia in which all Principal Investigators were young researchers (minimum of 2/maximum of 9 years post-PhD). A total Call pre-proposals were submitted to Call , of which 34 proposals were funded, corresponding to a success rate of 23.6%. Of the consortia funded, 11 contain Portuguese teams, 1 of the consortia is coordinated by a young Portuguese researcher and has the participation of another Portuguese team. The FCT participated in the three Calls total funding of approximately €1.37 million.
Projects Funded with Portuguese Participation
Call
• Analysis of the cellular mechanisms underlying the early response of the host to stress induced by Listeria infection. Project led in Portugal by Didier Cabanes (IBMC), with a total budget for three years of €100,000.
• Helicobacter pylori diversity in pathogenesis, antibiotic resistance, and evasion from natural and vaccine-induced immune responses. Project led in Portugal by José Carlos Machado (IPATIMUP), with a total budget for three years of €100,000.
• Global analysis of antisense regulatory mechanisms in Staphylococcus aureus. Project led in Portugal by Susana Domingos (ITQB), with a total budget for three years of €87,500.
• Characterization of host cell pathways altered by effectors of Brucella, Chlamydia, and Coxiella: identification of novel therapeutic targets. Project coordinated in Portugal by Jaime Mota (ITQB) with the participation of another national research team led by João Paulo Domingues (INSA), with a total budget of €200,000 over three years .
Call
• ADHRES-Signature Project. Project led in Portugal by Isabel Sá-Correia (IST), with a total budget for three years of €93,432.
• Pathogenomics of increased Clostridium difficile virulence. Project led in Portugal by Adriano Henriques (ITQB), with a total budget for three years of €181,800.
• Development, prevention, and early diagnostic detection of Clostridium difficile-associated pseudomembranous colitis—an interdisciplinary network. Project led in Portugal by Miguel Godinho Lifewizz Lda., with a total budget for three years of €142,000.
Call
• Parasite and host genetic diversity in Helicobacter infections. Project led in Portugal by José Carlos Machado (IPATIMUP), with a total budget for three years of €95,000.
• Large-scale screening of potential key factors involved in the commensalism/virulence transition of Enterococcus faecalis. Project led in Portugal by Maria de Fátima Silva Lopes (ITQB), with a total budget for three years of €86,435.
• A global RNAi approach to unravel eukaryotic host functions that modulate bacterial infections. Project led in Portugal by Céu Figueiredo (IPATIMUP), with a total budget for three years of €180,000.
• Spatio-temporal analysis of Listeria-host protein interactions. Project led in Portugal by Didier Cabanes (IBMC), with a total budget for three years of €112,962.
Awards
One of the objectives of PGM was to promote and encourage training in the genomics of microorganisms that cause disease in humans. In this context, between 2004 and 2012, six Calls were opened Calls reward the best doctoral theses in this area. Each selected student received a prize of €2,000 and was offered the opportunity to participate in an international conference with an oral presentation of their doctoral work. During this period, the PGM awarded prizes to 18 doctoral theses, three of which were by Portuguese researchers:
• João Paulo Gomes (2007): “Contribution to the understanding of biological differences among Chlamydia trachomatis serovars using genomics and transcriptomics.”
• Cristina Dias Rodrigues (2008): “Revealing Host Factors Important for Hepatocyte Infection by Plasmodium.”
• Alexandra Isabel Cardoso Nunes (2010): “Genomic and Transcriptomic Features of Chlamydia trachomatis: Tracking the Basis for the Ecological Success.”